DNE: A Deep Machine Learning-based Network Encoder that Uncovers Non-linear Gene-gene Functional Associations at Systems Level | |||||
|
|||||
Summary ^
Deep Network Encoder (DNE), a novel network reverse engineering method
developed in Hu Li's lab
that harness the power of autoencoder and demonstrated that it is
possible to uncover non-linear gene-gene associations learned
from high-dimensional omics data.
Genes do not act alone but are interdependent to each other to execute their functions.
These mutual interactions between genes are the basis for the non-linear nature of the
gene-gene functional associations in a biological network.
We realized that the network architectures of deep learning models have similar analogies
with real-world networks (e.g. biological networks) where information encoded in weights
connecting each nodes in learned models can enable us to decipher non-linear relations
between features (e.g. gene-gene associations) during learning processes.
That is, deep neural network models can be used as knowledge discovery platforms to
uncover non-linear relations between features in real world.
With this key insight, we developed Deep Network Encoder (DNE) to decipher non-linear
relations of features from high-dimensional data. We devised association scores where
non-linear association for a pair of genes can be inferred using weights of all possible
paths that connect these genes from input to output nodes. A biological network that
contained highly associated gene pairs will be reverse engineered.
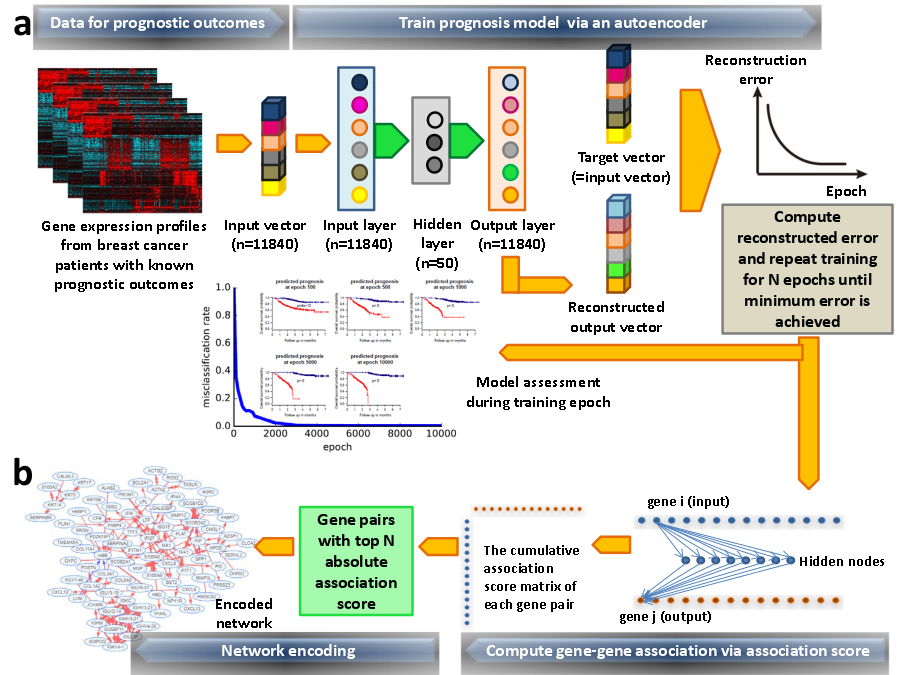
Overview of Deep Network Encoder (DNE) algorithm using breast cancer prognosis as an illustrative example. (a) Model building and training. Gene expression profiles of breast cancer patients with known prognostic outcomes were assigned as good and poor prognosis according to disease relapse-free survival (DRFS) that inform survival length of patients without cancer relapse. Models corresponding to good and poor prognosis respectively were bulit. Autoencoder algorithm was used to train the models, with input vector represents all genes present in transcriptomics data and the expression value of each gene corresponds to respective node at input layer. The dimensionality of output layer (i.e. number of nodes) is the same with input layer. The aim of autoencoder training is to reconstruct values from input layer at the output layer. The resulting output vector generated from output layer will be compared with input vector to compute error of reconstruction. The training process was repeated by updating weights connecting nodes (or neurons) from input layer to hidden layer and from hidden layer to output layer via backpropagation algorithm. Training process will come to halt when no further improvement on reconstruction error was achieved. (b) Network encoding via trained model. Weights connecting all nodes (neurons) from input to output layers in a trained autoencoder model were used to encode non-linear gene-gene associations using an association scoring scheme. Computed association scores for all gene pairs were given in an association score matrix where gene pairs with top 200 absolute scores were selected. Genes that occur multiple times in these top 200 gene pairs will served as "seeds" to agglomerate gene pairs into a network. Results ^
Explore gene association networks derived from autoencoder models for phenotype groups here. Download ^
Download the scripts
with sample dataset to run DNE on your local Linux system.
Extract the scripts and sample datasets into the same folder
and read the README.txt file to get started. Support ^
For support of DNE, please post to our web forum. Citation ^
Manuscript in preparation. |